Validation of PD-L1 AI model launches in six labs across the Netherlands
Labs in the Netherlands are joining together to launch an AI model validation study for PD-L1 scoring with a goal of multi-source domain adaptation.
Introduction
Programmed death-ligand 1 (PDL1) is a protein that, when attached to T cells, prevent the immune system from killing cancer cells. Hence, PDL1 is used as a biomarker for tumor immune escape, a sign of increasing cancer progression. PDL1 percentage is also a significant predictor for immunotherapy efficacy, whereby immune checkpoint inhibitors bind to PDL1 and block its ability to bind to T cells. This data is thus crucial for clinicians to prescribe the best course of treatment for cancer patients.
A team at Leiden University Medical Center (LUMC) created a deep learning algorithm, or AI model, for PD-L1 scoring in non-small cell lung cancer (NSCLC) histology with Aiforia Create. Having accomplished an accuracy rate equal to pathologists analysing the slides themselves, the team’s next goal is to develop a multi-source domain adaptation of the model, changing the future of NSCLC diagnosis.
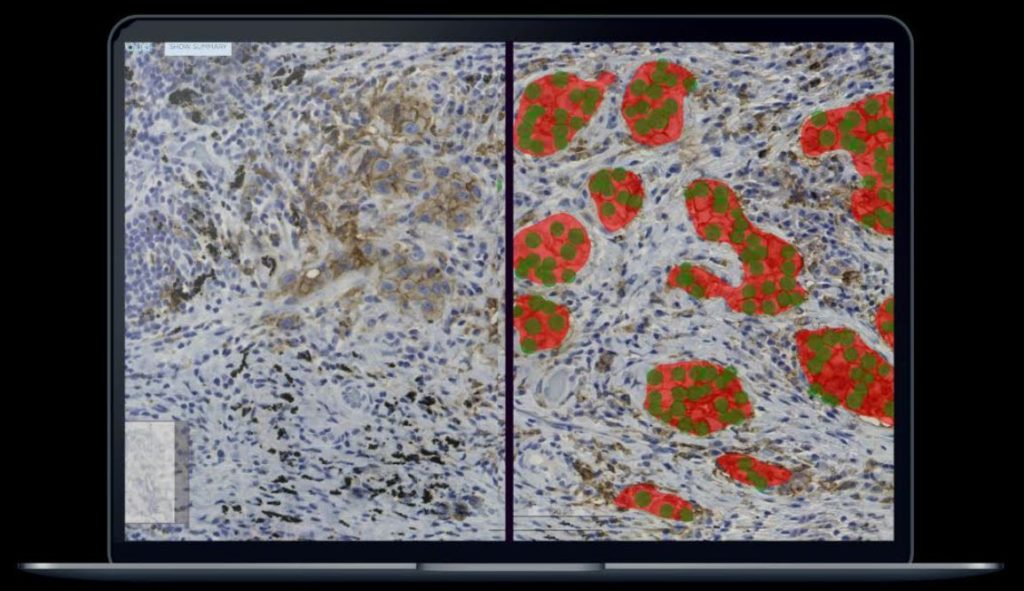
The AI model will be validated with preexisting setups in six different labs across the Netherlands – LUMC, EMC, Isala, HMC, NKI, and AMC. The labs will test the PDL1 scoring model with their own methods of sample preparation, testing for adaptation flexibility. In primary trials 50% of slides were scanned and shared with the LUMC project lead Liesbeth Hondelink. The project contains a mix of Philips and Hamamatsu scanners, 2 different antibodies, and has been divided into two setups:
Primary: Parallel algorithm update training process. Take LUMC algorithm as base and then re-train based on the new slides from each center.
Secondary: Sequential algorithm update training process. LUMC algorithm updated with Isala, and the resulting algorithm is then updated with EMC, etc.
Labs involved
LUMC: Leiden University Medical Center
EMC: Erasmus University Medical Center
Isala
HMC: Haaglanden Medisch Centrum Antoniushove
NKI: The Netherlands Cancer Institute
AMC: University of Amsterdam Academic Medical Center
Interview with Project Lead
Read about the team’s experience using Aiforia AI models through our interview with project coordinator Liesbeth Hondelink, MD, a PhD-candidate in the Department of Pathology at Leiden University Medical Center (LUMC), the Netherlands.
Tell us a bit about the validation project (what is the aim, who and how many institutions are involved, etc.)?
Liesbeth: The aim is to describe the steps required for multi-source domain adaptation for digital immunohistochemistry.
In this era of digital pathology, more and more artificial intelligence (AI) based applications will become available. These AI-models have the potential to improve diagnostic accuracy and save time for pathologists and researchers. However, most algorithms are not widely applicable to new workups at all, due to known differences between laboratories. A different scanner, stainer, antibody, microtome or laboratory protocol can result in a substantial drop in algorithm accuracy.
Variance in slide color and intensity is therefore high between laboratory workups, especially in immunohistochemistry. Only 6% of all published medical AI-algorithms are validated on an external dataset, a percentage that is expected to be even less in immunohistochemistry-based algorithms. Robust recommendations on how to start using an existing algorithm in a new pathology work-up are therefore mostly lacking.
In this study, we describe the steps required for generalizing an existing algorithm for PD-L1 assessment to a new workup via semi-supervised domain adaptation (adding training data from the new workup in order to become equally accurate in those circumstances). We will investigate which differences in workups are the most problematic for the AI, and require the most intensive re-training.
We have started a multi-center collaboration with 6 laboratories in the Netherlands.
How will the AI model be validated in each lab?
After domain adaptation on a ‘training set’, we will validate the algorithm performance against the pathologists scores for each participating laboratory in a separate ‘validation set’.
What is unique about the project?
It is the first multi-source domain adaptation project for digital immunohistochemistry. We hope to describe a step-by-step approach for domain adaptation, that can be easily implemented in any laboratory.
Why did you decide to use the Aiforia software for this?
We created the PD-L1 algorithm using Aiforia software, so it was logical to proceed with it.
What is the biggest benefit of the Aiforia software for this validation project?
Scalability and explainability. Aiforia uses ‘explainable AI’ as a cornerstone in the software, which makes it easy to use and interpret, even when you lack machine learning knowledge.
Find out more about AI for image analysis: www.aiforia.com